Article: Protocol for genetic load analysis in caribou using a modified genomic evolutionary rate profiling
Authors: Rebecca S. Taylor, Micheline Manseau, Peng Liu, Paul J. Wilson
Highlights
- Procedure for using an automated pipeline for whole-genome multi-species alignment
- Instructions for running a modified genomic evolutionary rate profiling program
- Guidance on interpretation of evolutionary conservation scores
- Steps to use a custom script to extract derived alleles from a VCF file using results
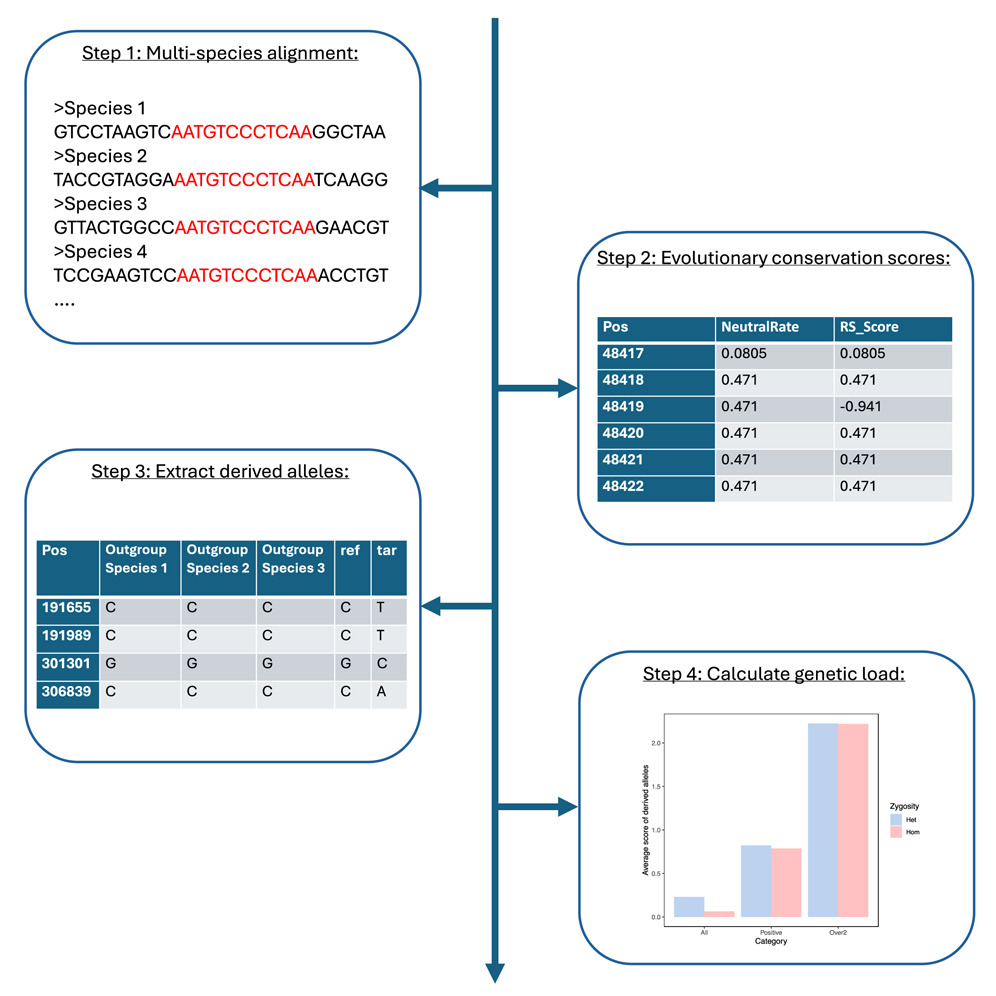
Summary
Here, we present a protocol to analyze genetic load in caribou (Rangifer tarandus) using a modified version of the genomic evolutionary rate profiling (GERP) program. We describe steps for multi-species alignment including automation of input file production and generating evolutionary conservation scores. We detail procedures for streamlining the extraction of sites of interest from a variant calling format (VCF) file as known derived alleles using three outgroup species to enable the measurement of genetic load.
For complete details on the use and execution of this protocol, please refer to Taylor et al. 2024.
Click here to access the full publication.




