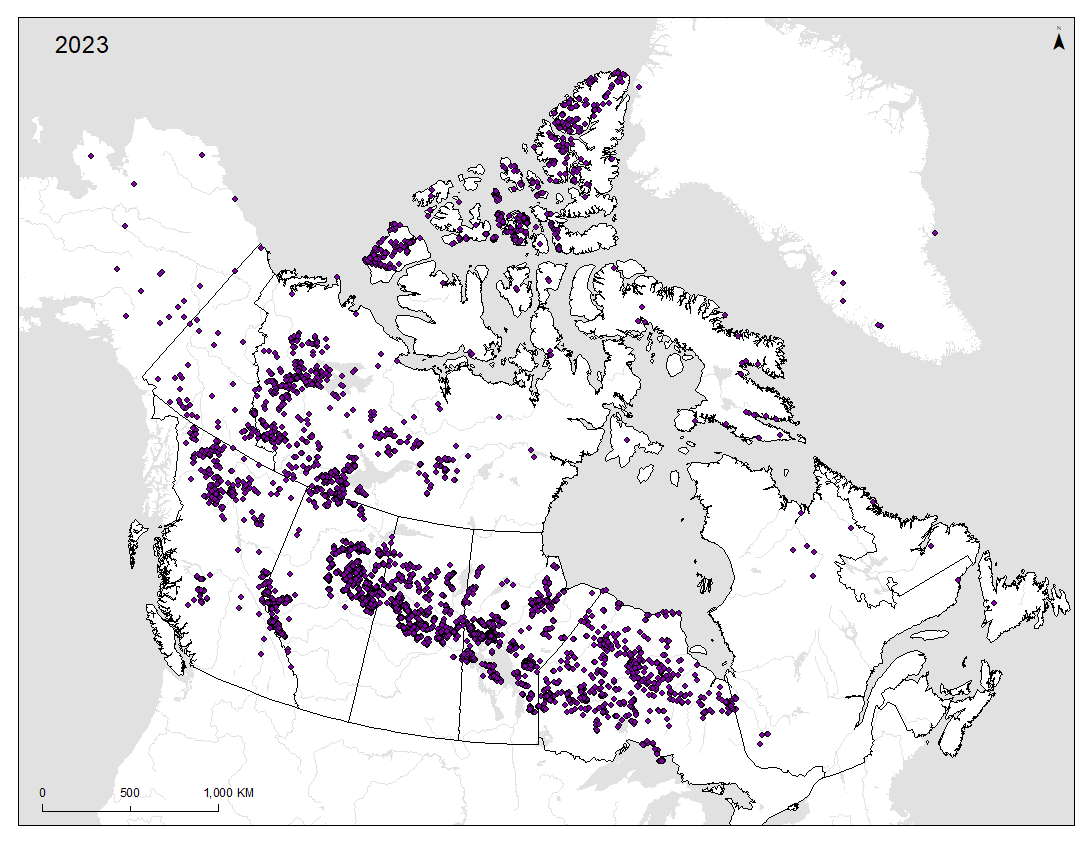
EcoGenomics is a research program that employs non-invasive fecal collection to monitor Canadian caribou. This longitudinal project considers ecological and evolutionary applications relevant to the conservation and management of this vulnerable species.
While focused on caribou, this program has research and monitoring implications for a wide range of species such as moose, deer and wolves.
The overarching objective of the EcoGenomics program is to create a long-term, Canada-wide caribou monitoring framework. This research utilizes standardized genomic methods and open-access data sharing tools. Achieving long-term caribou monitoring will substantially increase:
- The capacity of government, industry, and Indigenous communities to participate in wildlife monitoring;
- Transparency, reliability, and cross compatibility of wildlife monitoring data; and
- Opportunities for collaboration across sectors in the monitoring and conservation of at-risk wildlife.
Our Story
Dr. Paul Wilson of Trent University and Dr. Micheline Manseau of Environment & Climate Change Canada (ECCC), collectively have more than 60 years of experience in wildlife genetic research. The two researchers initiated caribou genetic research in 2004 after discovering that high quality and quantities of DNA could be extracted from the mucosal coat of caribou pellets. The extraction of DNA permits identification of individual caribou by means of genetic markers. Since their early investigations, Wilson and Manseau have overseen the genetic profiling of over 40,000 caribou pellets nationwide. In collaboration with long-term research partners, this data has allowed for the unique identification of individual caribou.
 The EcoGenomics Team is currently building upon previous research, together with collaborations and partnerships to harness:
The EcoGenomics Team is currently building upon previous research, together with collaborations and partnerships to harness:
- The power of large-scale DNA sequencing technology, metagenomics, remote sensing, advances in bioinformatics and artificial intelligence, and the sequencing of the entire genetic complement of an individual animal’s DNA; and
- The expansion of an already extensive database of fecal and DNA specimens and associated genetic data from across the country dating back 20 years into a standardized open-access platform which maximizes the ability to monitor caribou over time.
This national-scale, collaborative project is supported by a Genome Canada Genomic Applications Partnership Program (GAPP) grant entitled, “Caribou genomics: A national non-invasive monitoring approach for an iconic and model species-at-risk.”
Wildlife Monitoring and Fecal DNA

Species Identification
Individual Identification
Whole-Genome Sequencing
Diet Characterization
Pregnancy Detection
Stress Response
Internal Microbiome
Pathogen Detection
The application of individual-specific profiles from non-invasively collected fecal pellets provides the ability to:
- Estimate species’ population size and trends over time;
- Estimate and map species’ density and other population parameters;
- Reconstruct species’ familial networks and pedigrees;
- Delineate species’ population structure to assess connectivity at demographic and population scales;
- Model the influence of the environment on species’ population density, diversity and connectivity;
- Model the influence of adaptive genes through Machine and Deep Learning algorithms; and
- Provide phylogenomic evolutionary analyses to delineate ecotypes and Designatable Units (DUs) under the Committee on the Status of Endangered Wildlife in Canada (COSEWIC).
Beyond high-quality genomic profiling, additional information about an animal’s health can be obtained from fecal material including:
 Profile targeted adaptive genes and even whole genomes;
Profile targeted adaptive genes and even whole genomes;- Metabarcoding to determine diet, the microbiome and pathogens; and
- Hormone analysis to determine pregnancy rates and stress levels.
Our Partners & Funders




























